Virus Therapeutics
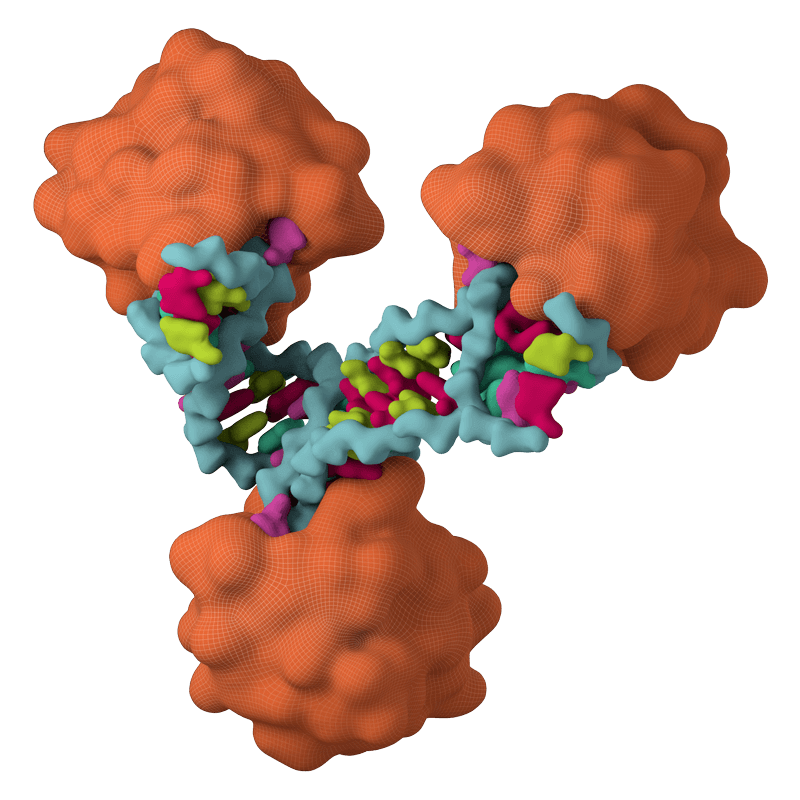
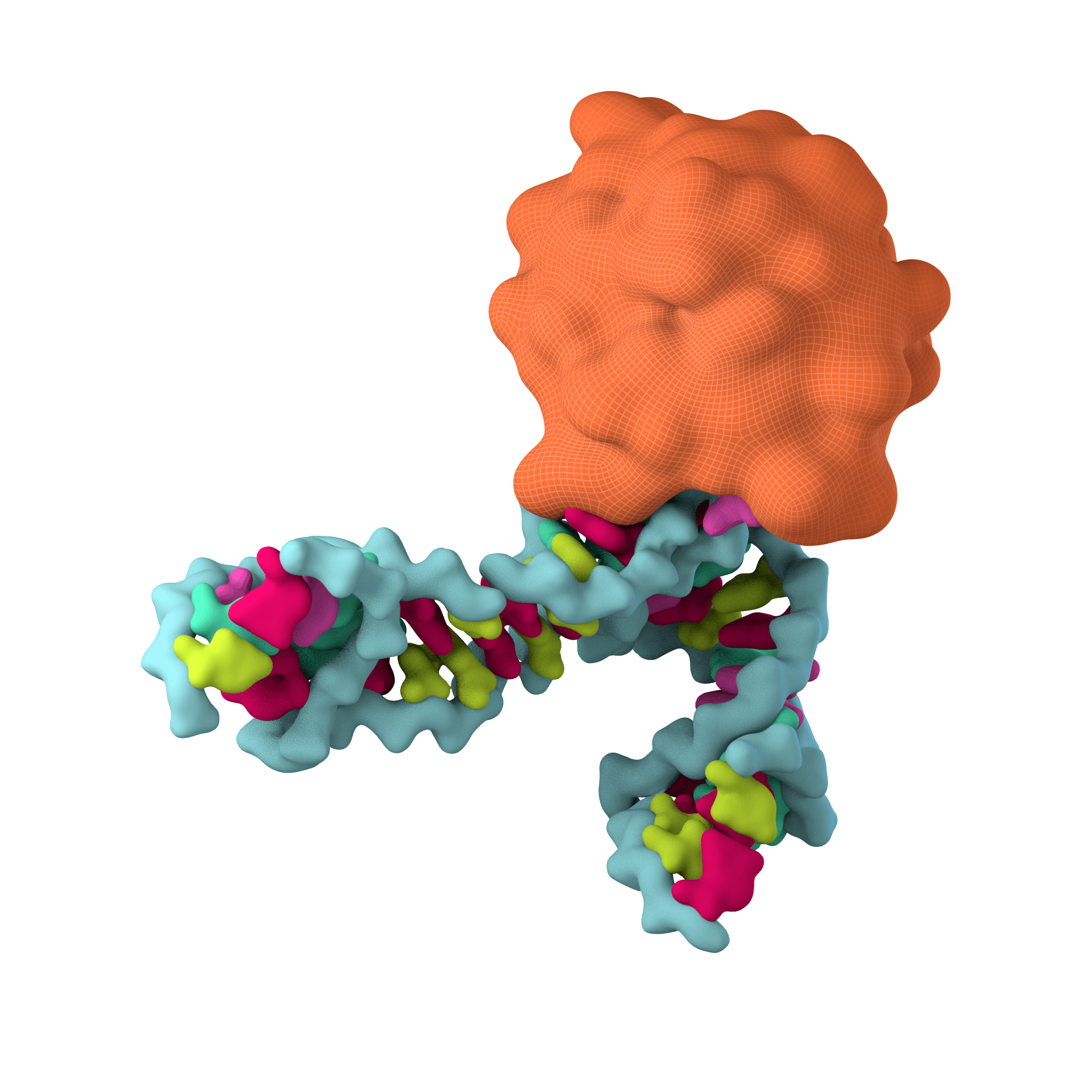
Viruses are obligate parasites that rely on host machinery to enable replication. The tenOever lab, in part, focuses on identifying products that can interfere with this biology. This program focuses on two distinct tools. First, an RNA binding protein found in archaea extremophiles and second, defective interfering genomes, both of which can be utilized to inhibit virus propagation.
CRISPR-Cas systems are a central platform in combating virus infection. In an effort to mine for additional defenses in archaea, we focused on the transcriptomics of virally infected Methanococcus maripaludis. Bulk RNA sequencing of M. maripaludis, infected with a novel member of the myovirus family, identified ~250 genes that were significantly upregulated by more than one order of magnitude. To circumvent issues of genome annotation and identifiable orthologues of M. maripaludis, we focused specifically on DNA and RNA binding proteins that were upregulated in response to infection. To this end, we synthesized 30 genes as codon optimized open reading frames for expression in mammalian cells and initially tested them against seven distinct virus challenge models to ascertain whether any showed antiviral activity. This screen identified a subset of genes that could either enhance or inhibit virus replication.
Bulge-Helix-Bulge (BHB) protein: Showed selective inhibition of positive strand RNA viruses (poliovirus, PV; Langat virus, LV). This gene product (Operon 585-1) is a member of the dsRNA-specific nucleases related to the RNase III family, which has been implicated in both the Type II CRISPR-Cas system and in the processing biology of RNAi. Given these results, we chose to clone members of this gene family from representative kingdoms to assess whether this activity was conserved. These data demonstrated that an inherent property of each of these proteins was to engage RNA hairpins commonly found in the genomes of positive stranded viral RNA and to induce their cleavage. Together, these data may reflect the evolutionary history of antiviral systems. This nuclease family may have been repurposed during the process of eukaryogenesis.
Unlike prokaryotes, which are infected almost exclusively by DNA-based viruses, eukaryotic viruses include both DNA and RNA viruses with a larger proportion being the latter. It has been proposed this phenomenon occurred because nuclear entry provided a new barrier for DNA viruses while the expansion of cellular membranes created an ideal environment for RNA viruses. In this scenario, early eukaryotes may have evolved to use their RNAse III arsenal to engage incoming RNA viruses and cleave any hairpins formed during their replicative process. This action alone would have provided some level of positive selection but the more significant advancement would later come with the capacity to utilize the RNA cleavage products by coupling them to an otherwise nonspecific nuclease such as the Argonaute family members. This would represent the earliest form of RNAi. In fact, based on homology, it appears as though antiviral RNAi arose before the split between animals and plants and that this biology was independently duplicated thereafter and used for developmental purposes in the form of miRNAs. It is thus interesting that vertebrates encode two RNAseIII nucleases in our genomes, Dicer and Drosha, of which the later shows greater homology to the overall family and is the only one of the two that demonstrates any antiviral activity. This hypothesis not only provides an understanding for how antiviral pathways may form over evolutionary time, but it also explains why RNAse III nucleases show consistent antiviral activity across all life.
Operon 169 (O160) of M. Maripaludis: This putative RNA binding protein was induced more than 100 fold and was found to potently block influenza A virus infection when introduced into mammalian cells. The closest orthologue of this gene in humans is the ribosomal protein called RPL30. In a manner similar to what we did for the RNAse III family members, we chose to synthesize codon-optimized components of the RPL30 family of ribosomal proteins, which are conserved in all three domains of life, to assess whether expression could disrupt influenza A virus replication. Members of this ribosomal protein from Plasmodium to Xenopus were expressed in human lung alveolar cells and were infected with the 2004 pandemic H1N1 influenza A virus (IAV). These data revealed that only members of the archaea family induced viral inhibition which we later determined was the result of blocking production of NS2, an alternative splice product of the virus required for replication. In addition to determining that expression of O169 could inhibit every strain of influenza virus we tried (n=8), we also noted that it had no impact on host splicing or the mammalian cell suggesting it represents a universal inhibitor of IAV infections.